Distill TotalSegmentator with Monai and Keras 3
TotalSegmentator is a tool for medical segmentation of most major anatomical structures. If you are starting a new project and you need quick segmentations, this tool is great with a permissive license! But it comes with a some costs:
- You dont controll the data. Usually in ML projects most improvments come from identifing cases your model is wrong and adding their type to training, but with TotalSegmentator you cannot as their dataset is paritally private and finetuing this project doesnt look easy.
- TotalSegmentator learned to segment many structures and you might need only a few. You might be able to use a mutch smaller faster model depending on your task.
Instead of using TotalSegmentaor as is, you can use it to create “weak labels” on your data. This could be a great start for creating initial sementation masks for your annotation team.
The following post will show how to create weak labels for the BTCV dataset with Totatlsegmentator and evaluate training on them compared to training on the real labels. I use the Monai framework and as an experiment wraped it in keras 3 with a pytoch backend. The code to reproduce is in my github
BTCV Data
50 abdomen CT scans (30 train, 20 test) with train segmentation labels for spleen, right/left kidneys, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, portal/splenic vein, pancreas, and right/left adrenal glands.
A typical slice with its segmentations looks like:
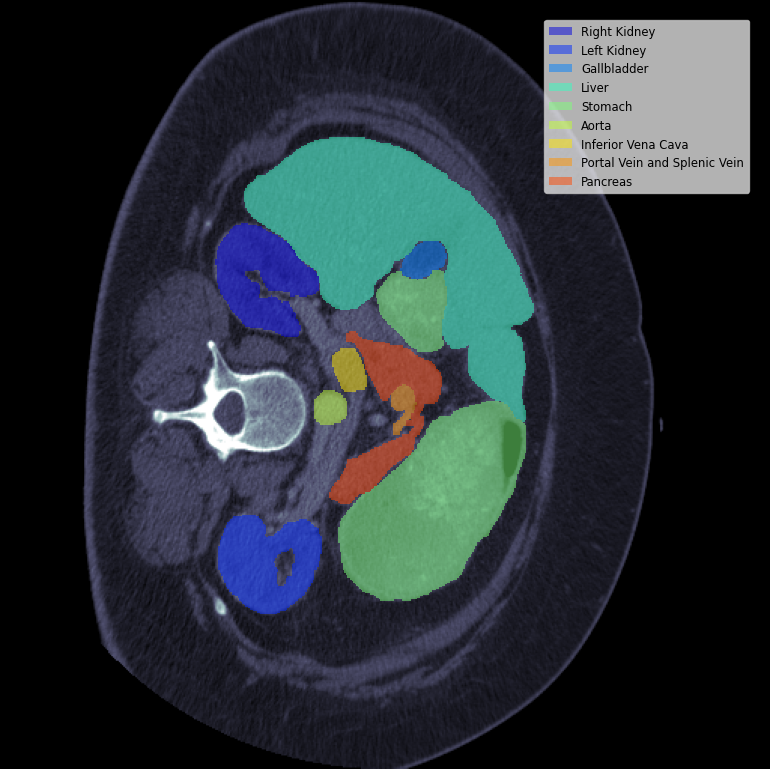
Becuase there are only labels for the train set thats what im going to use for train + validation. The validation images are the same used in many monai examples such as this one.
Create Weak Labels
Creating “weak labels” in this case means using TotalSegmentator to generate segmentation masks for the entire dataset and saving them in the same format as the original labels.
for nii_file in nii_files:
output_folder = Path(args.output_path) / (nii_file.stem[:-4])
totalsegmentator(
nii_file,
output_folder,
fast=True,
roi_subset=[
"spleen",
"kidney_right",
"kidney_left",
"gallbladder",
"liver",
"stomach",
"aorta",
"inferior_vena_cava",
"portal_vein_and_splenic_vein",
"pancreas",
"adrenal_gland_right",
"adrenal_gland_left",
"esophagus",
],
)
masks = []
for i, f in enumerate(label_files_order, start=1):
m = nib.load(output_folder / f).get_fdata()
m = m * i
masks.append(m)
mask = np.sum(masks, axis=0)
# Load the original NIfTI file to get the header and affine
original_nii = nib.load(nii_file)
new_nii = nib.Nifti1Image(mask.astype(np.int32), original_nii.affine, original_nii.header)
new_nii.header.set_data_dtype(np.int32)
new_nii.to_filename(labels_folder / f"{nii_file.name.replace('img','label')}")
Training
First, create a PyTorch dataset. Since the BTCV dataset is small, I’m loading it entirely into memory and applying the slow deterministic_transform functions once.
One of the deterministic transforms is resizing all the CTs to the same pixel spacing, which is time-consuming and shouldn’t be done during the training loop. If your dataset is too large to fit into memory, create a processing.py script and save the preprocessed images before training. The training loop should only contain random transformations that cannot be applied beforehand.
class BTCVDataset(torch.utils.data.Dataset):
def __init__(self, data, deterministic_transform, transform):
self.transform = transform
self.data = [deterministic_transform(d) for d in tqdm(data, desc="Loading data")]
def __len__(self):
return len(self.data)
def __getitem__(self, index):
return self.transform(self.data[index])
Check out the transformations and dataloader in train.py
SwinUNETR Keras Model
To create a Monai model and use it in Keras, first create the torch model:
model = SwinUNETR(
img_size=patch_size,
in_channels=1,
out_channels=14,
feature_size=48,
use_checkpoint=True,
)
To wrap a torch model in a Keras layer, I need to use keras.layers.TorchModuleWrapper. However, since it wouldn’t work without overriding the compute_output_spec, I created a new CustomTorchWrapper. The output_channels parameter represents the number of labels.
class CustomTorchWrapper(keras.layers.TorchModuleWrapper):
def compute_output_spec(self, inputs_spec, ouptut_channels=14):
h, w, d = inputs_spec.shape[2:] # Get spatial dimensions from input
output_shape = (None, ouptut_channels, h, w, d)
return keras.KerasTensor(shape=output_shape, dtype="float32")
During training, I want to evaluate the model with Monai’s sliding_window_inference function. To do that, I subclass keras.Model and override the test_step function.
class SlidingWindowValidationModel(keras.models.Model):
def __init__(self, patch_size, sliding_window_batch_size=4, *args, **kwargs):
super().__init__(*args, **kwargs)
self.patch_size = patch_size
self.sliding_window_batch_size = sliding_window_batch_size
def test_step(self, data):
x, y, sample_weight = data_adapter_utils.unpack_x_y_sample_weight(data)
x, y = x.to(get_device()), y.to(get_device())
y_pred = sliding_window_inference(
x, self.patch_size, self.sliding_window_batch_size, partial(self, training=False)
)
return self.compute_metrics(x, y, y_pred, sample_weight)
And finally, I use the Keras functional API to create the Keras version of the model:
inputs = keras.layers.Input(shape=(1, *patch_size))
x = CustomTorchWrapper(model)(inputs)
k_model = SlidingWindowValidationModel(patch_size, 4, inputs, x)
Adjusting Losses
To use Monai’s DiceCELoss loss, I need to make some adjustments. Monai’s loss expects the order of ypred, ytrue to be loss(ypred, ytrue), but during Keras’s fit function, the loss is called with loss(ytrue, ypred). To solve this I have a wrapper loss:
class DiceCELossKeras(torch.nn.Module):
def __init__(self, to_onehot_y, softmax, *args, **kwargs):
super().__init__(*args, **kwargs)
self.l = DiceCELoss(to_onehot_y=to_onehot_y, softmax=softmax)
def forward(self, y_true, y_pred):
return self.l(y_pred, y_true)
Metrics
I want to use Monai’s DiceMetric. I’ll create a Keras Metric wrapper for it. To speed up training, I want to apply this metric only during the validation stage. This is a reasonable decision since during training the model sees only (96,96,96) patches, while during validation I’m evaluating the whole image.
class MonaiDiceMetricKeras(keras.metrics.Metric):
def __init__(self, include_background=True, reduction="mean", get_not_nans=False, *args, **kwargs):
super().__init__(name="monai_dice", *args, **kwargs)
self.m = DiceMetric(include_background=include_background, reduction=reduction, get_not_nans=get_not_nans)
self.post_label = AsDiscrete(to_onehot=14)
self.post_pred = AsDiscrete(argmax=True, to_onehot=14)
def reset_state(self):
self.m.reset()
def update_state(self, y_true, y_preds, sample_weight=None):
if torch.is_grad_enabled() or y_true.device == torch.device("meta"):
# dont compute for train set or keras build stage
return
y_true_list = decollate_batch(y_true)
y_true_convert = [self.post_label(val_label_tensor) for val_label_tensor in y_true_list]
y_preds_list = decollate_batch(y_preds)
y_preds_convert = [self.post_pred(val_pred_tensor) for val_pred_tensor in y_preds_list]
self.m(y_pred=y_preds_convert, y=y_true_convert)
def result(self):
if self.m.get_buffer() is None:
return 0.0
return self.m.aggregate().item()
And finally, train the model using Keras fit
model.compile(
optimizer=keras.optimizers.AdamW(learning_rate=1e-4, weight_decay=1e-5),
loss=DiceCELossKeras(to_onehot_y=True, softmax=True),
metrics=[MonaiDiceMetricKeras()],
run_eagerly=False,
)
model.fit(
train_dl,
validation_data=val_dl,
epochs=1500,
validation_freq=20,
)
Distillation
Finally, I can run my training script twice: once with the training dataset using the original BTCV labels, and once with the training set using the weak labels. The validation uses the original labels in both cases.
Here are the validation results for both cases:
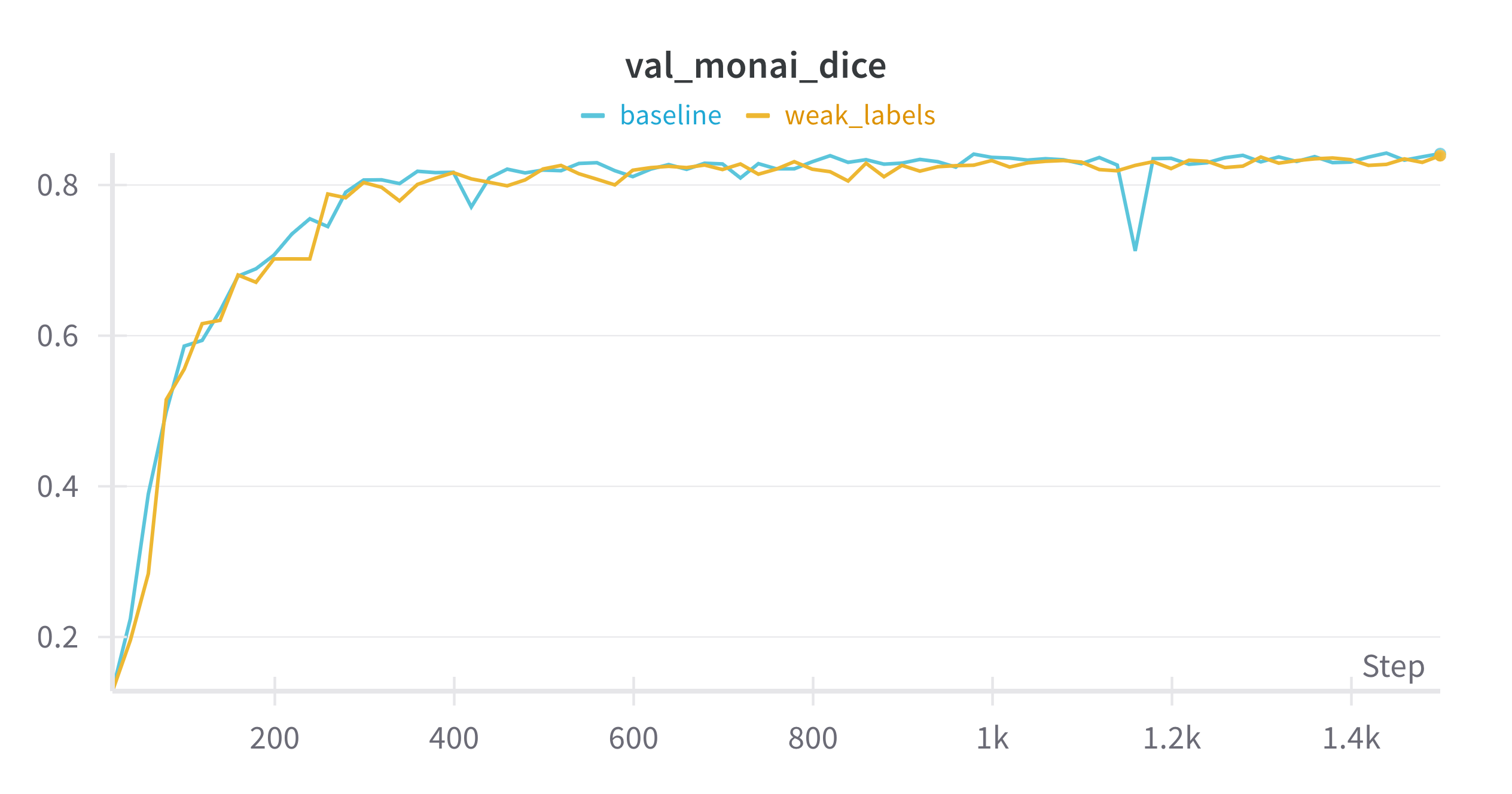
Not bad! Results are almost the same as training on real labels. This means that training on weak labels is as good as training on real labels, and if this were a real project, I could start off without labeling data, just using TotalSegmentator as my annotator.
Of course there are some caveats: this is a very small dataset with an even smaller validation set, and TotalSegmentator was likely trained on BTCV data and is really good on it. It would be interesting to run this experiment on data TotalSegmentator has never seen.
These results are encouraging, and next time I start a new medical AI project, instead of starting with time-consuming annotations from medical experts, I can start labeling data with TotalSegmentator and iterate.